Cholesky paths with umxPath
This page will introduce writing Cholesky structures using umxPath to facilitate twin modeling with umxRAM.
It is not finished, and for now (and perhaps for sometime), I’d recommend you read this page instead on the awesome umxACE function
# ====================
# = Cholesky example =
# ====================
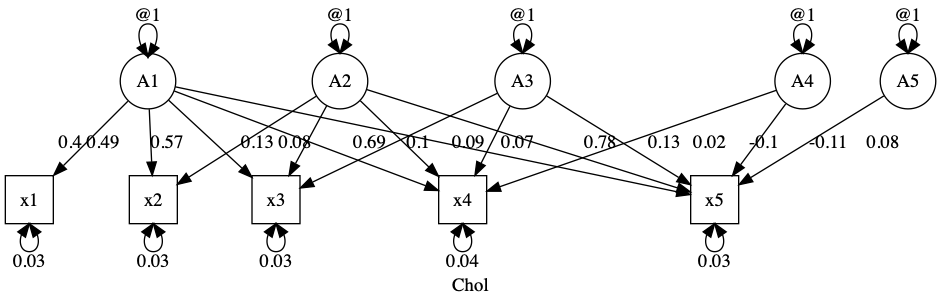
latents = paste0("A", 1:5)
manifests = names(demoOneFactor)
myData = mxData(cov(demoOneFactor), type = "cov", numObs = 500)
m1 = umxRAM("Chol", data = myData,
umxPath(Cholesky = latents, to = manifests),
umxPath(var = manifests),
umxPath(var = latents, fixedAt = 1.0)
)
We can see that this fits “perfectly” - it’s a saturated solution, with some caveats (see umx::umxACEv to learn more about these):
umxSummary(m1)
What about a 1-factor solution?
A 1 factor Cholesky is just a general factor, but let’s use it as an example:
manifests = names(demoOneFactor)
m2 = umxRAM("Chol", data = demoOneFactor, type = "cov",
umxPath(Cholesky = "A1", to = manifests),
umxPath(var = manifests),
umxPath(var = "A1", fixedAt = 1)
)
plot(m2)
We could also drop the paths from m1:
m3 = umxModify(m1, regex = "^A[2-5]")
tip: use parameters(m1, pattern = "^A[2-5]") to see what paths this expression matches and drops.
Is a one-factor model significantly worse than the extravagant saturated five Cholesky-factor solution?
umxCompare(m1, m2)
| Model | EP | Δ -2LL | Δ df | p | AIC | Δ AIC | Compare with Model |
|---|---|---|---|---|---|---|---|
| Chol | 20 | 9.994993 | 0 | ||||
| Chol | 10 | 7.3987999 | 10 | 0.687 | -2.606207 | -12.6012001 | Chol |
No: fit is not-significantly worse.
A multi-group twin model
Twin models have models for each class of twins – MZ DZ, perhaps differentiated by sex – and these differ in the genetic paths they set.
umx includes built-in twin models, including umxACE. Here, we’ll build a twin model completely by hand in umx from scratch.
NOTE: umx now includes umxTwinMaker which makes specifying novel multivariate twin models MUCH more straight-foward.
First let’s build an ace model from scratch:
data(twinData)
tmpTwin = twinData
names(tmpTwin)
# "fam", "age", "zyg", "part", "wt1", "wt2", "ht1", "ht2", "htwt1", "htwt2", "bmi1", "bmi2"
# Set zygosity to a factor
labList = c("MZFF", "MZMM", "DZFF", "DZMM", "DZOS")
tmpTwin$zyg = factor(tmpTwin$zyg, levels = 1:5, labels = labList)
# Pick the variables
selDVs = c("bmi1", "bmi2") # nb: Can also give base name, (i.e., "bmi") AND set suffix.
# the function will then make the varnames for each twin using this:
# for example. "VarSuffix1" "VarSuffix2"
mzData = tmpTwin[tmpTwin$zyg %in% "MZFF", selDVs]
dzData = tmpTwin[tmpTwin$zyg %in% "DZFF", selDVs]
latentA = paste0(c("A1"), "_T", 1:2)
latentC = paste0(c("C1"), "_T", 1:2)
latentE = paste0(c("E1"), "_T", 1:2)
latents = c(latentA, latentC, latentE)
mz = umxRAM("mz", data = mxData(mzData, type = "raw"),
umxPath(v1m0 = latents),
umxPath(v.m. = selDVs),
# twin 1
umxPath(Cholesky = "A1_T1", to = "bmi1"),
umxPath(Cholesky = "C1_T1", to = "bmi1"),
umxPath(Cholesky = "E1_T1", to = "bmi1"),
# twin 2
umxPath(Cholesky = "A1_T2", to = "bmi2"),
umxPath(Cholesky = "C1_T2", to = "bmi2"),
umxPath(Cholesky = "E1_T2", to = "bmi2"),
# A C E links
umxPath("A1_T1", with = "A1_T2", fixedAt = 1),
umxPath("C1_T1", with = "C1_T2", fixedAt = 1),
umxPath("E1_T1", with = "E1_T2", fixedAt = 0)
)
dz = mxModel(mz, name= "dz",
mxData(dzData, type = "raw"),
umxPath("A1_T1", with = "A1_T2", fixedAt = .5)
)
m1 = mxModel(mz, dz, mxFitFunctionMultigroup(c("mz", "dz")))
parameters(mz)
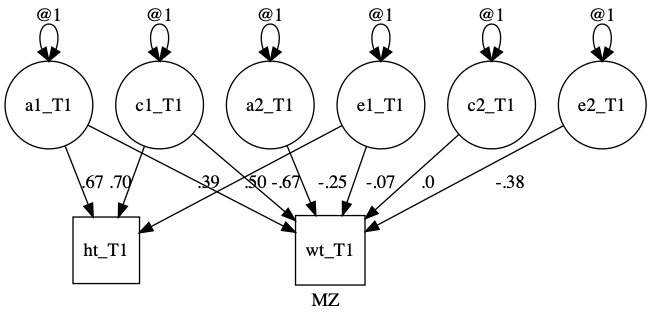
plot(dz, fixed= TRUE)
umxTwinMaker
Now let’s do that in umxTwinMaker. Here we only have to draw the model for one person.
First let’s assemble some data
data(twinData)
tmp = umx_make_twin_data_nice(data=twinData, sep="", zygosity="zygosity", numbering=1:2)
tmp = umx_scale_wide_twin_data(varsToScale= c("wt", "ht"), sep= "_T", data= tmp)
mzData = subset(tmp, zygosity %in% c("MZFF", "MZMM"))
dzData = subset(tmp, zygosity %in% c("DZFF", "DZMM"))
Now, I’ll make a bivariate ACE twin model.
1. Define paths for one person:
latents = paste0(rep(c("a", "c", "e"), each = 2), 1:2)
paths = c(
umxPath(v1m0 = latents),
umxPath(mean = c("wt", "ht")),
umxPath(fromEach = c("a1", 'c1', "e1"), to = c("ht", "wt")),
umxPath(c("a2", 'c2', "e2"), to = "wt")
)
2. Make a twin model based on the paths for one person
m1 = umxTwinMaker("myACE", paths, mzData = mzData, dzData= dzData)
plot(m1, std= TRUE, means= FALSE)
Here’s the same thing using umxACE
m2 = umxACE(selDVs= c("wt", "ht"), mzData = mzData, dzData=dzData, sep="_T")
plot(m2)